Heading 1
Heading 2
Heading 3
Heading 4
Heading 5
Heading 6
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
Block quote
Ordered list
- Item 1
- Item 2
- Item 3
Unordered list
- Item A
- Item B
- Item C
Bold text
Emphasis
Superscript
Subscript
About This Simulation
Embark on a mission to identify the metabolic pathway that produces an antimalarial compound in a rare plant. Can you begin the process of creating a novel antimalarial drug?
Learning Objectives
- Understand how a gene can be involved in the synthesis of a compound
- Understand how Next Generation Sequencing technology can be used to measure gene expression
- Use BLAST and phylogenetic analysis to screen candidate genes
About This Simulation
Lab Techniques
- BLAST
- RNA-seq
Related Standards
- Alignment pending
- Alignment pending
- Alignment pending
Learn More About This Simulation
This simulation replaces the previous “Plant Transcriptomics” simulation.
What if you could engineer a metabolic pathway from a rare plant into another organism and produce an antimalarial compound at large scale? In this simulation, you will learn how to identify the genes that encode specific enzymes. You will use a method called RNA-seq to quantify the transcription of different genes. Use this information to narrow down the candidate genes by using bioinformatics tools.
Find a novel antimalarial compound
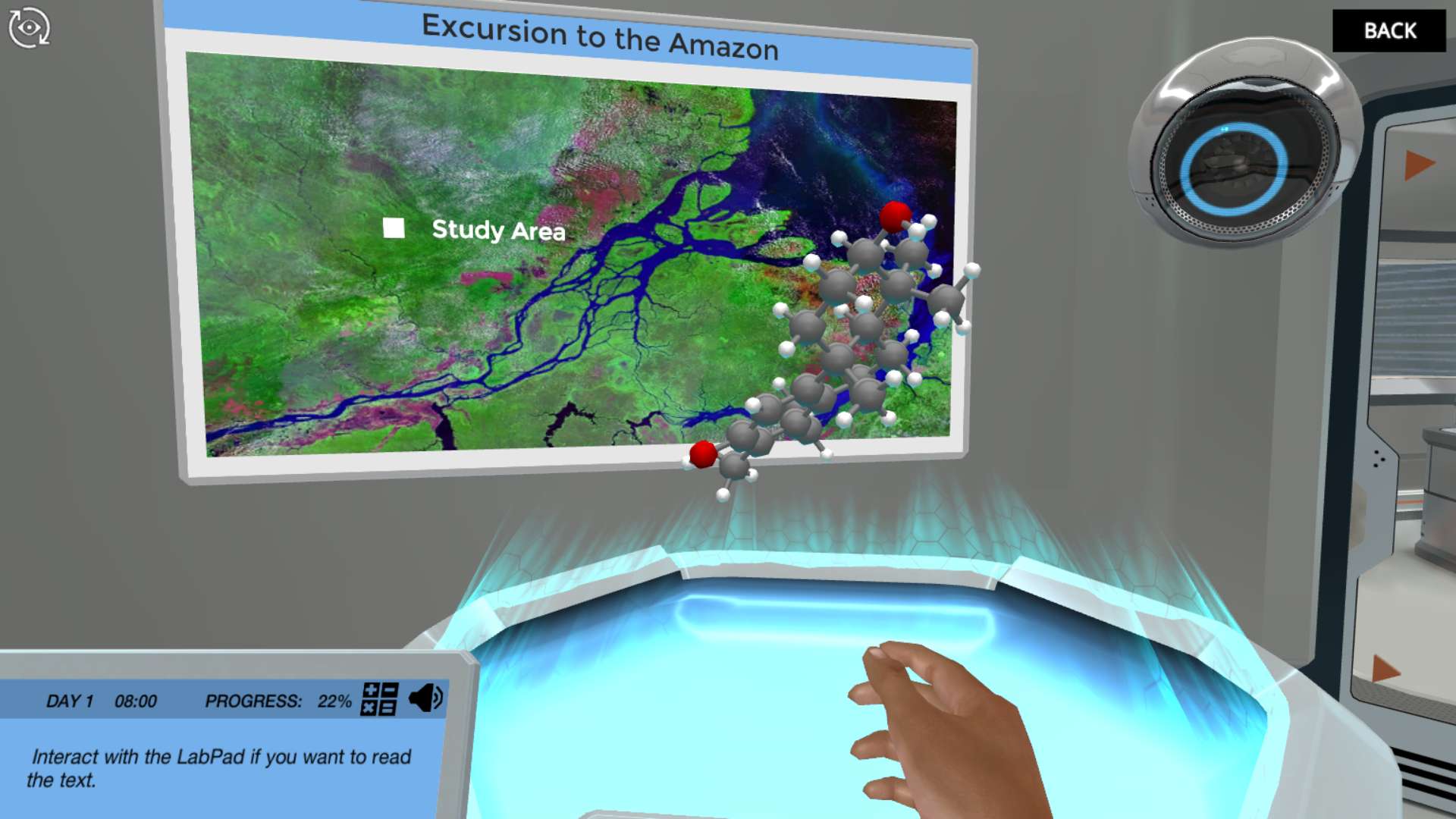
Your mission starts with analyzing a plant sample from the Amazon rainforest. A group of local tribal people use this rare plant to cure malaria. A phytochemist identifies the antimalarial compound. Your task is to identify the enzymes responsible for the production of this potential drug.
Learn how molecular compounds are produced and extract the RNA
Your next task will be to identify an enzymatic pathway that converts the precursor molecule into the antimalarial component. If you want to produce this compound at large quantities, you will need to transform the enzymatic pathway into another organism. To do so, you must identify the genes encoding the enzymes. All the information you need is contained within the composition of mRNA molecules in different plant tissues.
Learn about the structural difference of mRNA and DNA and perform RNA-seq
The key to the functions of mRNA and DNA is their structures. While the lab assistant is reverse transcribing your mRNA sample into cDNA, you will be learning about the structure of these two amazing molecules. Your next task is then to perform Next Generation Sequencing (NGS) of your cDNA sample. You will sequence the cDNA of different plant tissues to determine the sequence and expression of the target enzymes. An animation will show you what is going on inside the flow cell enabling you to see how millions of different sequences can be read in parallel.
Analyze your results using phylogenetic analysis and BLAST
Like a true detective, you will analyze the huge amount of NGS reads and determine which enzyme is most likely involved in the biosynthesis of the antimalarial compound. You will learn how relatedness of DNA sequences can be used to identify unknown enzymes and learn the basics of sequence comparisons.
Will you be able to identify the enzymes and produce a novel drug that can save millions of people?
For Science Programs Providing a Learning Advantage
Boost STEM Pass Rates
Boost Learning with Fun
75% of students show high engagement and improved grades with Labster
Discover Simulations That Match Your Syllabus
Easily bolster your learning objectives with relevant, interactive content
Place Students in the Shoes of Real Scientists
Practice a lab procedure or visualize theory through narrative-driven scenarios


FAQs
Find answers to frequently asked questions.
Heading 1
Heading 2
Heading 3
Heading 4
Heading 5
Heading 6
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
Block quote
Ordered list
- Item 1
- Item 2
- Item 3
Unordered list
- Item A
- Item B
- Item C
Bold text
Emphasis
Superscript
Subscript
A Labster virtual lab is an interactive, multimedia assignment that students access right from their computers. Many Labster virtual labs prepare students for success in college by introducing foundational knowledge using multimedia visualizations that make it easier to understand complex concepts. Other Labster virtual labs prepare learners for careers in STEM labs by giving them realistic practice on lab techniques and procedures.
Labster’s virtual lab simulations are created by scientists and designed to maximize engagement and interactivity. Unlike watching a video or reading a textbook, Labster virtual labs are interactive. To make progress, students must think critically and solve a real-world problem. We believe that learning by doing makes STEM stick.
Yes, Labster is compatible with all major LMS (Learning Management Systems) including Blackboard, Canvas, D2L, Moodle, and many others. Students can access Labster like any other assignment. If your institution does not choose an LMS integration, students will log into Labster’s Course Manager once they have an account created. Your institution will decide which is the best access method.
Labster is available for purchase by instructors, faculty, and administrators at education institutions. Purchasing our starter package, Labster Explorer, can be done using a credit card if you are located in the USA, Canada, or Mexico. If you are outside of North America or are choosing a higher plan, please speak with a Labster sales representative. Compare plans.
Labster supports a wide range of STEM courses at the high school, college, and university level across fields in biology, chemistry, physics, and health sciences. You can identify topics for your courses by searching our Content Catalog.











.png?fm=jpg&w=450&h=400)





